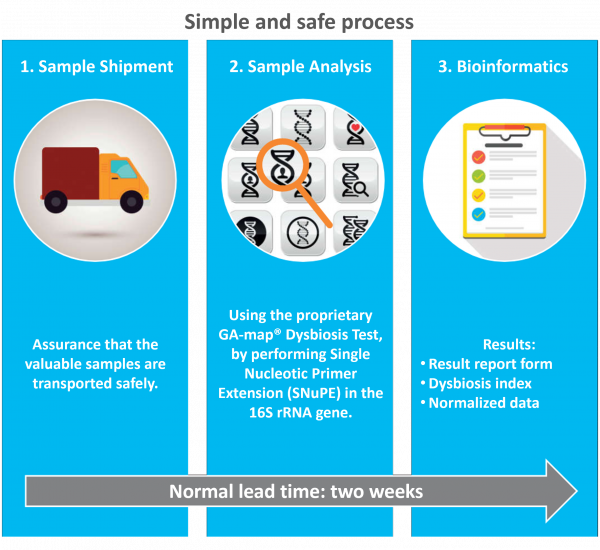
Report
Advanced bacterial analysis is presented in an easy-to-read report form. In addition, normalized data for all markers are provided for research use.
GA-map® Technology ![]()
Patented GA-map® technology for dysbiosis testing, using the Single Nucleotic Primer Extension (SNuPE) methodology, on the 16S rRNA gene, to identify predetermined bacteria at different taxonomic levels. The bacteria were specifically selected to separate a normal healthy microbiome profile from a dysbiotic profile.